Competing stems?
In preparation for a manuscript describing how the sxy gene is regulated in H. influenzae, we want to illustrate predictions of how sxy mRNA folds into stems and loops. A very useful program called Mfold can generate loads of pictures illustrating predicted secondary structure (ie. 2-dimensional representation of the stems and loops predicted to form in an RNA molecule). In addition to these predictions, we have experimental analysis of sxy mRNA secondary structure. Ultimately we want to compare and contrast Mfold's thermodynamic predictions with the experimental data of sxy mRNA folding.
However, we have a problem. Because RNA molecules are dynamic and can often fold into many alternate secondary structures that are all thermodynamically equally (or almost equally) stable, Mfold provides multiple predictions for a single RNA sequence. Comparing the data with predicted structures is all well and fine at positions where we have data, unfortunately the structural data is patchy, thus we lack experimental insight into some regions of the sxy molecule. Consequently we are left with the problem of deciding which Mfold prediction is “best” for these experimental blind spots.
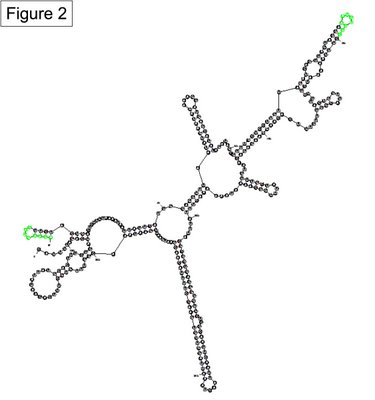
Experiments confirm the prediction that the 5’ end of sxy mRNA folds into what we call Stem 1A and Stem 1B. What we don’t know is whether Stem 1 also has a third region (Stem 1Z), or whether an adjacent Stem 4 forms from some of the same sequence. Figure 1 shows two alternate Mfold predictions, Structure 1 with Stem 1Z and Structure 2 with Stem 4 (the two structures differ in sequence length, but that difference is not important for this discussion). The bases that contribute to Stems 1Z or 4 are highlighted green to show that the two stems are mutually exclusive.
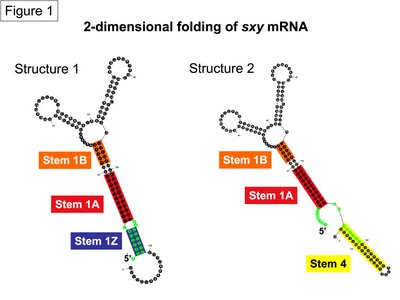
In the absence of experimental data for the green sequences that form stems 1Z or 4, I have tried to estimate the relative probability of sxy forming either stem by comparing multiple Mfold predicitons. I asked Mfold to predict the best structures for the 340 nucleotide sxy RNA molecule used in the experimental structural analysis. It returned 25 predictions with a Gibbs free energy (delta G) between -75.5 and -69.2. Of these predictions, both stems showed up in multiple structures across the full spectrum of this free energy range, indicating that neither stem contributes to a more thermodynamically favourable strucutre. However, Stem 4 (including variants that have longer and shorter Stem 4s with different loops at the end) formed in 8/25 structure while Stem 1Z formed in only 4/25 structures. The real surprise was that 14/25 structures looked nothing like our favourite model of sxy folding presented here (Figure 1). In these alternate structures, the green sequences are as diametrically opposed as possible on the large RNA molecule (an example is in Figure 2). These alternate structures do not immediately appear to agree with our genetic evidence of sxy mRNA folding, so we can ignore them for now. However, counting the frequency of stem formation in Mfold predictions has got us no closer to deciding on whether to draw sxy mRNA folding with Stem 1Z or Stem 4.