I am currently conducting what I think to be a cool experiment to turn the E. coli lac promoter into a Sxy-dependent (or possibly Sxy-enhanced) promoter. Earlier this year I received a very useful clone, pASKAsxy, from a group in Japan; it carries the E. coli sxy gene under strong LacI repression. Using this clone I discovered that Sxy induces E. coli competence genes, but not genes with normal CRP-N sites. Now I want to test whether it is the TGCGA motif of CRP-S sites that single out CRP-S promoters for activation by Sxy. In other words, does having the TGCGA motif make a promoter Sxy-dependent?
I have mutated the CRP site in the classic lac promoter (Plac) to change it from the CRP-N motif (TGTGA) to the CRP-S motif (TGCGA), which I call “Plac-S”. My hypothesis is that Sxy will strongly induce Plac-S, but not regular Plac (possible results are discussed below).
But first, a technical problem. Plac and Plac-S are each carried on the pSU20 vector that confers resistance to chloramphenicol. pSU20 uses the p15 origin of replication and is compatible with the pASKAsxy ColE1 origin, however pASKAsxy also confers chloramphenicol resistance. To remedy this antibiotic incompatibility I have excised Plac and Plac-S from pSU20 while at the same time removing Plac from pSU40, a close relative of pSU20 that confers kanamycin resistance. Today I cloned the pSU20-derived Plac and Plac-S into pSU40. Next I will clone both pASKAsxy and pSU40-Plac concurrently into DH5a.
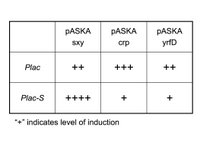
I can’t do a simple +/- sxy experiment as I have done in the past. The problem is that both sxy and Plac are LacI repressed, thus in the sxy- state (ie. no IPTG), Plac can’t be activated anyway. Thus, IPTG will always have to be present to detect differences in Plac induction levels. To control for sxy activity I will also clone pASKAcrp and pASKAyrfD with pSU40-Plac; the former clone is expected to stimulate Plac but not Plac-S, while the latter (which encodes the E. coli comA homologue) is not expected to activate either Plac or Plac-S (see Table). I will quantify Plac activity using real-time PCR directed at lacZ-alpha subunit mRNA. DH5a lacks lacZ-alpha, so the signal will only come from plasmids.

3 Comments:
This sounds really interesting and maybe you could talk me through the experiment at some point?
Where are the cloning diagrams? I was expecting some pretty pictures showing the logic of your experiments....no first born for you!!
These sounds very similar to the findings printed by Camron and Redfielod in NAR journal.
I am actually working on nutritional competence and yrfD is one of the genes involved in the process. The presence of CRP-S in front of the yrfD and its transcriptional dependance on sxy(yccR) in E. coli is very helpful to support my theory that dsDNA is utilized by E coli as a nutrient through the similar machinery involved in DNA uptake (homologs in Hi and Ng). That data was also published in J. of Bact. last year.
Post a Comment
<< Home